
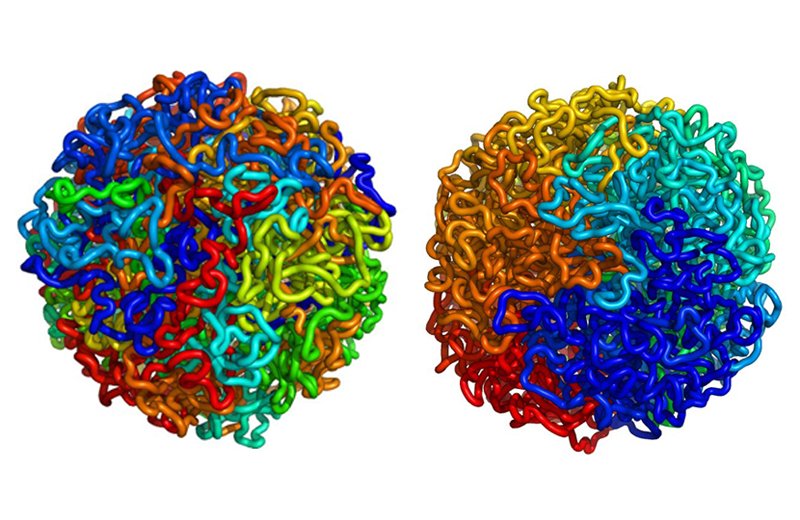
MIT chemists have developed a groundbreaking artificial intelligence (AI) technique to predict the three-dimensional (3D) structure of the genome, a discovery that could revolutionize the study of gene expression.
Led by Associate Professor Bin Zhang, the team created ChromoGen, a generative AI model capable of analyzing DNA sequences and predicting chromatin structures in single cells within minutes—far faster than traditional experimental methods like Hi-C, which can take a week per cell.
“Our goal was to predict the 3D genome structure from the DNA sequence,” Zhang explained. “Now that we can do that, it really opens up a lot of interesting opportunities.”
ChromoGen combines deep learning and generative AI to process DNA sequences and chromatin accessibility data, producing thousands of possible chromatin conformations. Since DNA structures are highly variable, the model accounts for multiple possible formations, enhancing its accuracy.
Once trained, the AI model significantly outperforms existing techniques in speed and scalability. “Whereas you might spend six months running experiments to get a few dozen structures, you can generate a thousand structures in 20 minutes on a single GPU,” said MIT graduate student Greg Schuette, a lead author of the study.
The researchers tested their model on over 2,000 DNA sequences, finding its predictions closely matched experimental data. The model can also generalize to different cell types, making it a valuable tool for studying how chromatin structure influences gene expression and disease-related mutations.
Published in Science Advances, this research was supported by the National Institutes of Health. The team has made their model and data publicly available, paving the way for further exploration of genome organization and its impact on cellular function.



Leave a comment